Y-Compare/Match Help
YCompare displays the kits that match the one specified in either the Compare or Matches tools.
It lists all the Marker allele values and highlights any differences from the 1st (top) kit specified.

Columns:
- User ID - The Identification number given to the kit when created. Click on this link to see details for this kit.
- EKA Surname - Surname of the Earliest Known Ancestor (EKA) on the direct patralineal line (Y-DNA test)
- EKA Birthplace - EKA's place of birth
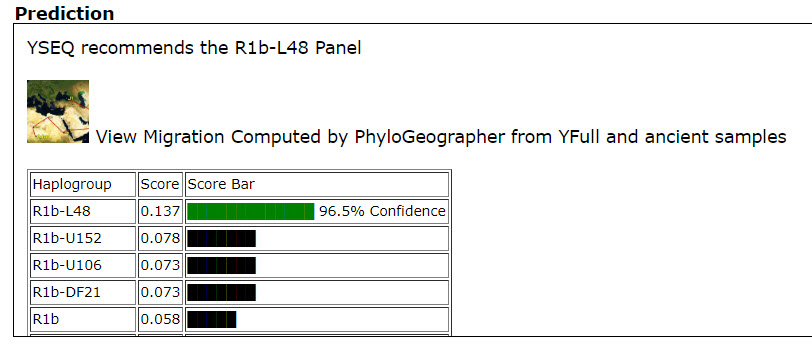
- Haplogroup - Haplogroup entered by user
- Genetic Distance (4)
- DIST 25
- DIST 37
- DIST 67
- DIST 111
- Marker Values - Short Tandem Repeat (STR) marker values (alelles) that represent your Haplotype (the set of DNA alelle values; not to be confused with Haplogroup). The figure above only shows part of the chart; it goes out to 111 markers. The color codes of the DYS headers reflects the range of markers tested by that kit.
| Color | Marker Range |
|---|---|
| 1-12 | |
| 13-25 | |
| 26-37 | |
| 38-67 | |
| 68-111 | |
| Fast Changing |
Y-DNA Genetic Distance
When talking about two or more Y-Chromosome STR haplotypes, Genetic Distance (GD) is the total number of differences, or mutations, between two sets of results.
The lower the number, the closer you are related.
In general, it is found by summing the differences between each STR marker.
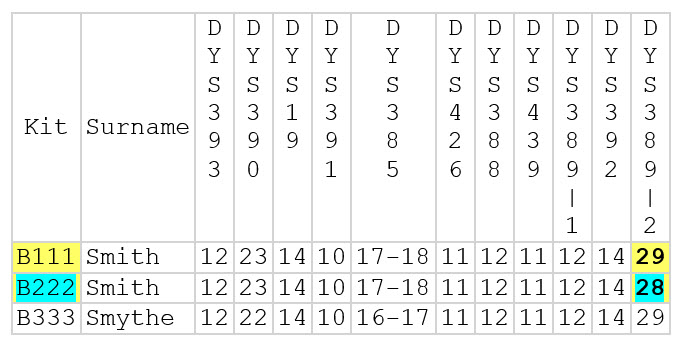
For example (see chart below), kit B111 and B222 have allele values of 29 and 28 respectively at DYS389-2. This is a difference of 1 {29-28= 1}. Because this is the only difference in their Y DNA12 haplotype, their genetic distance is 1.
It has nothing to do with Generations; just math!
In July 2017, FTDNA introduced changes in how the genetic distance is calculated for some multi-valued markers (eg., DYS385, DYS464). Roberta Estes has an excellent blog entry about this change here.